We process personal information and data on users of our website, through the use of cookies and other technologies, to deliver our services, personalize advertising, and to analyze website activity. We may share certain information on our users with our advertising and analytics partners. For additional details, please refer to our Privacy Policy.
By clicking “AGREE” below, you agree to our Terms of Use and Privacy Policy , as well as our personal data processing and cookie practices as described therein. You also consent to the transfer of your data to our servers in the United States, where data protection laws may be different from those in your country.
Data
C0-expression networks
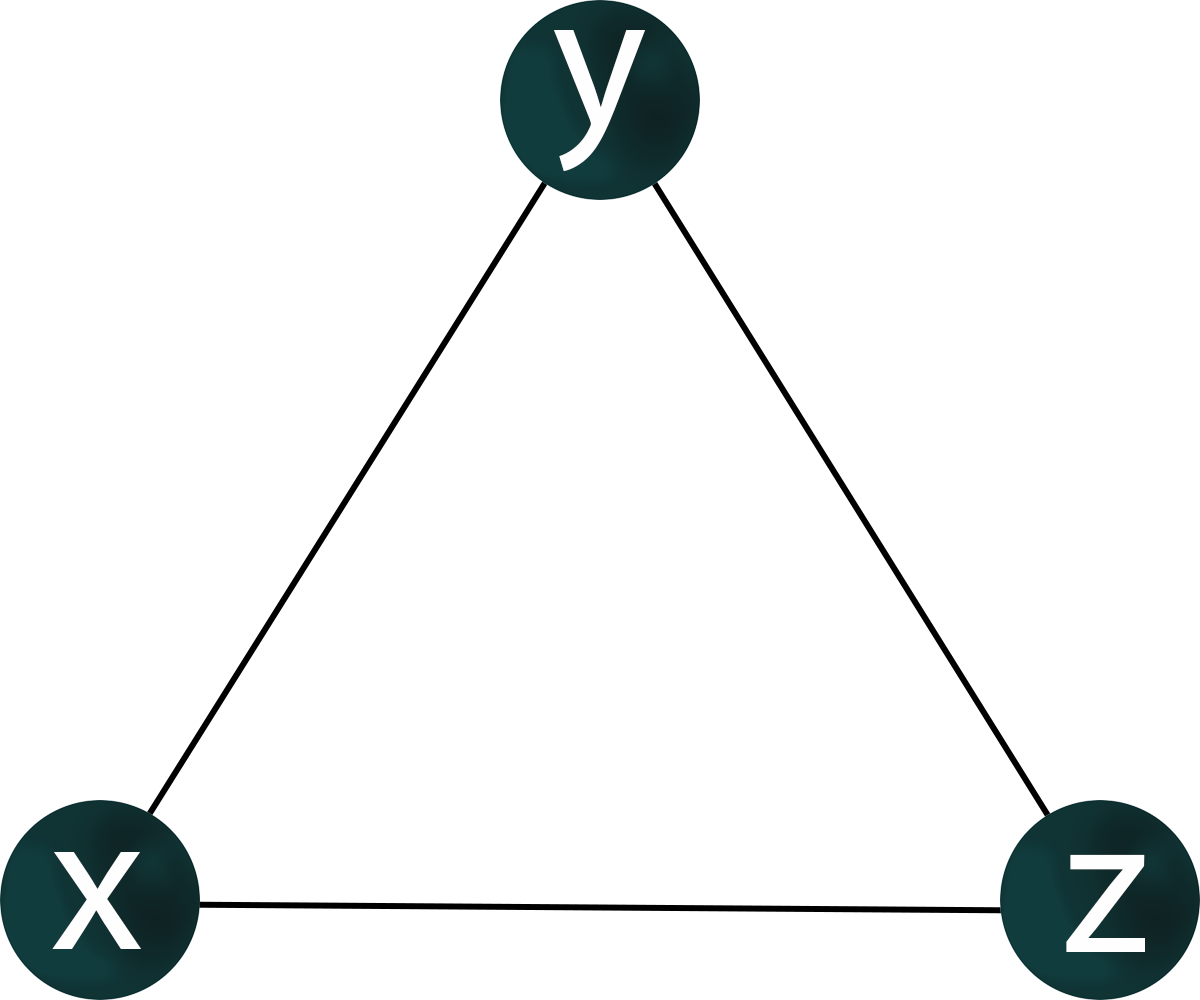
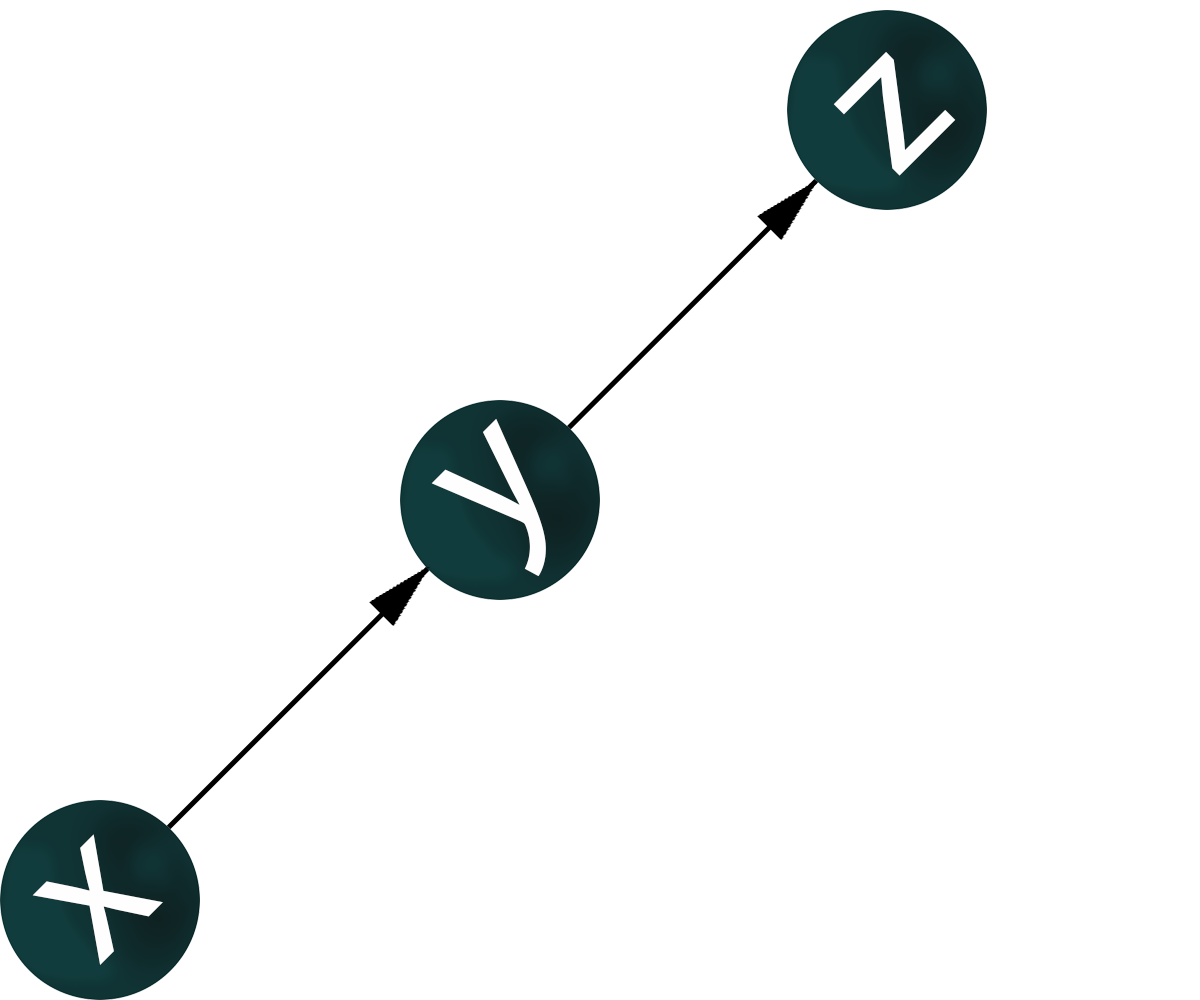
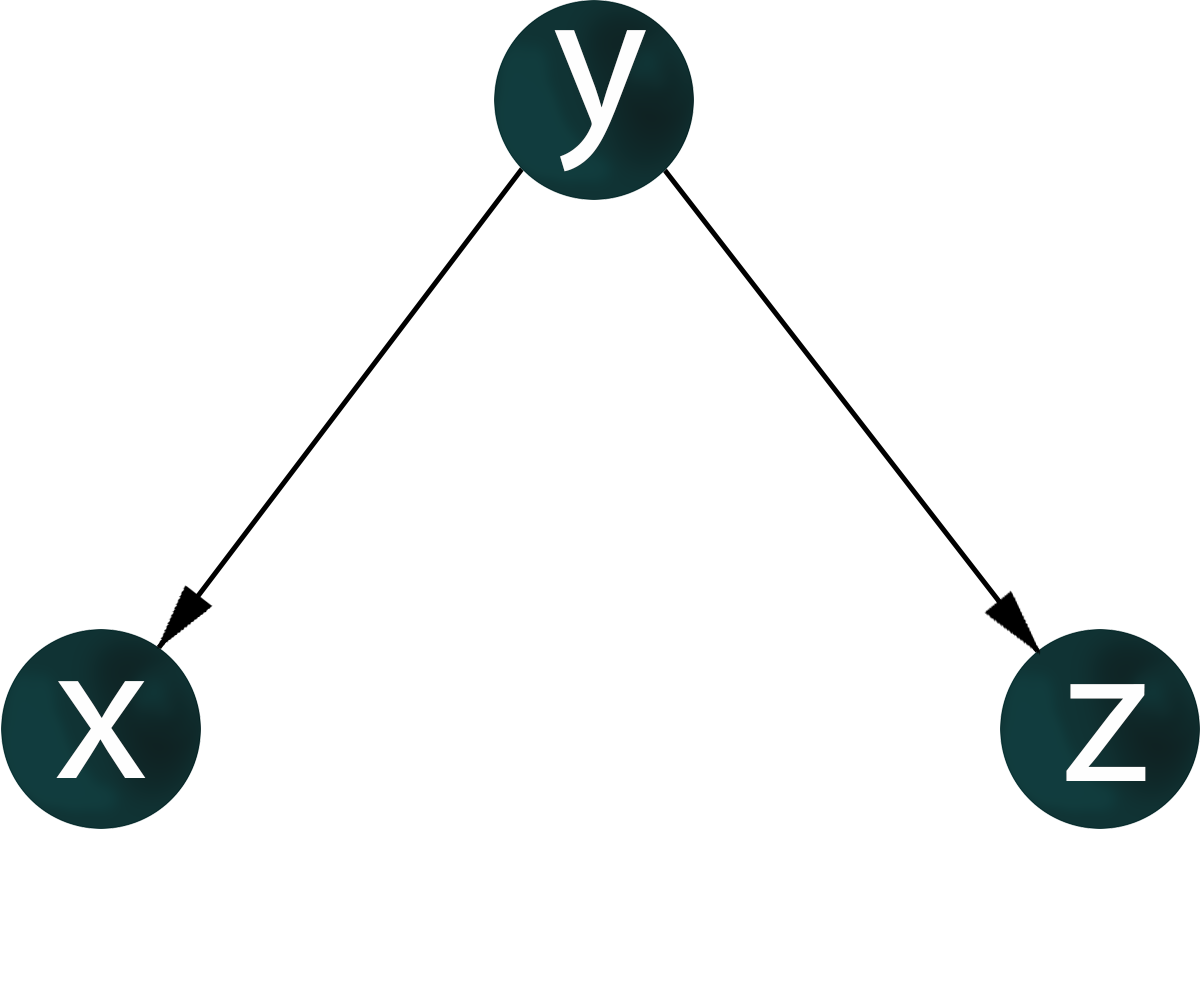
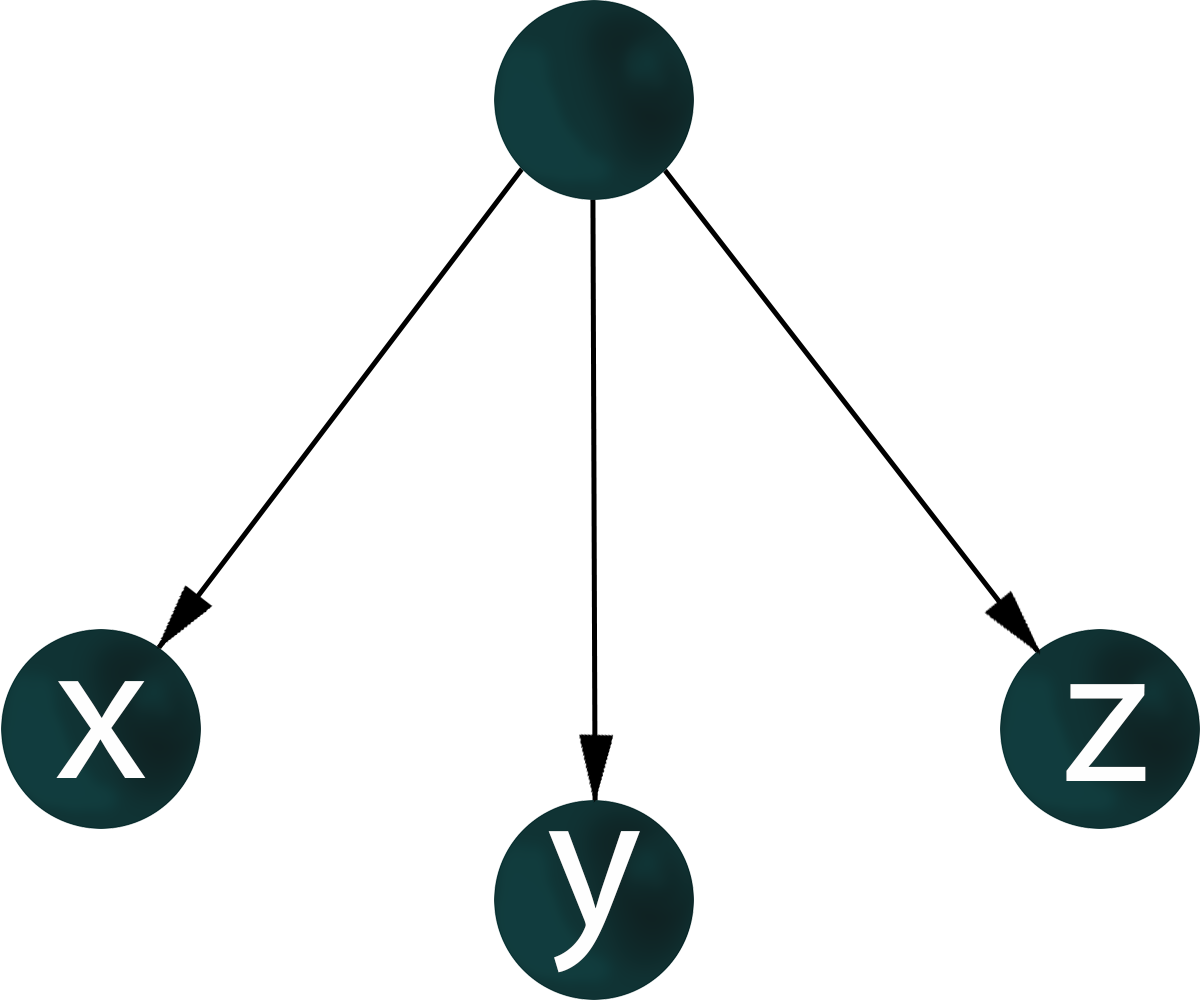
If genes x, y, z are co-expressed, nothing is known about regulatory relationships. For the co-expression of genes x, y, z, it is possible to construct a graph where the vertices corresponding to these genes have pairwisw connections.
A gene-regulatory network is an oriented graph in which for a pair of genes it is known which gene is a transcription factor – activator or repressor, and which gene is regulated.
A co-expression network is an undirected graph in which edges mean relationship between expressions of genes, corresponding to vertices. Thus, the expression of two genes may be correlated, but one gene does not necessarily regulate the other.
The coexpression network and the gene regulatory network may have different structures.
A co-expression network is an undirected graph in which edges mean relationship between expressions of genes, corresponding to vertices. Thus, the expression of two genes may be correlated, but one gene does not necessarily regulate the other.
The coexpression network and the gene regulatory network may have different structures.
There is no complete information about all interactions of genes in the gene regulatory network, so we can only approximate the structure of the network by various methods.
For a given structure of co-expression network of genes x, y, z, there may be many variants of the topology of the gene regulatory network.

